Virtual Laboratory for Plant Breeding Github Repository
This repository contains data analysis tools developed within Virtual Laboratory for Plant Breeding Project.
RNA-seq analysis pipelines
Small RNA pipelines have beed developed within Virtual Laboratory for Plant Breeding projects. The pipelines make use of mostly R and Python. The diagram that has been used to build these pipelines:
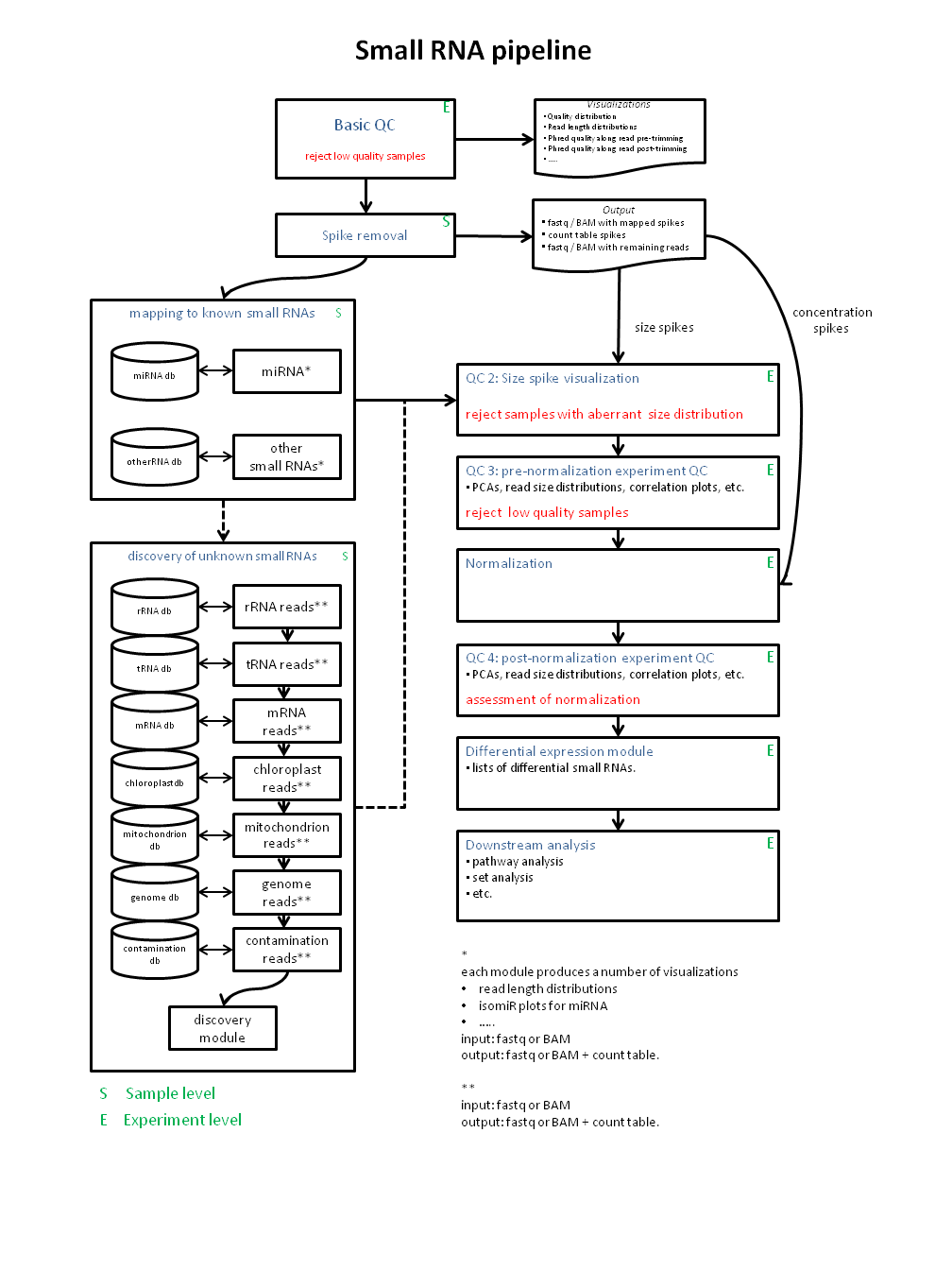
Currently the Basic QC, Mapping, Spike removal & visualization, normalization and pre- and post visualization are ready to use.
Every pipeline is run in separate environment. By encapsulating most of the dependecies together with analysis code.
Different levels of dependencies:
system level packages
python
- Python version
- Python moudule versions
both can be fixed with use of python virtual environment
R
- R version
- R packages version the system level R version (first one in the path) is used
Right versions of R packages are ensured with use of Packrat. That alows for building whole R environment with rigth R packages.
Packrat environment works at the level of directory.
# example basicQC analysis directory# with all necessary Packrat related files and directories.├── basicQC.htm # html file with QC report├── basicQC.md # markdown file genereated by knitr├── basicQC.Rmd # Rmarkdown defining the QC report├── figure # plots│ ├── A.design.plot-1.png│ ├── C3plot-1.png│ ├── D1plot-1.png│ └── D2plot-1.png├── get_experiment_fq.py # script that imports read data form sff files into fq files├── packrat # dir with R environment related files│ ├── init.R # script initializing Packrat environment│ ├── lib #│ ├── lib-ext # R library│ ├── lib-R #│ ├── packrat.lock # log of all the package versions used by this environment│ ├── packrat.opts # packrat options│ └── src├── raw # fastq files generated from sff files│ ├── N406T.fastq│ ├── N540T.fastq│ ├── N583T.fastq│ └── sknas.fastq├── README.md # project readme file├── seqdesign.txt # experiment design file└── Snakefile # pipeline defined for Snakemake
Pipelines and tools developed at the University of Amsterdam:
-
faradr
R package used across the pipelines.
-
basicQC
Quality control of raw reads in context of the experiment.
-
rnaxcount
Alignment and counting fo small RNA species.
-
rnaxqc
Quality control of small RNA counts in the context of the experiment.
-
sRNA-norm
Normalisation for counts based on spike-in counts.
Glossary:
- Packrat
- Packrat is a dependency management system for R. Read more.
- Snakemake
- Python module and library for building workflows. Project's aim is to reduce the complexity of creating workflows by providing a fast and comfortable execution environment, together with a clean and modern domain specific specification language (DSL) in python style. Read more.
- virtualenv
- A tool to create isolated Python environments. See Read more.